
Tools to run the tests Automatic execution script for setup_daq Analysis Tools
RGANG tool introduction Command line execution on multiple hosts Tools on LXSHARE
rename partition node selection
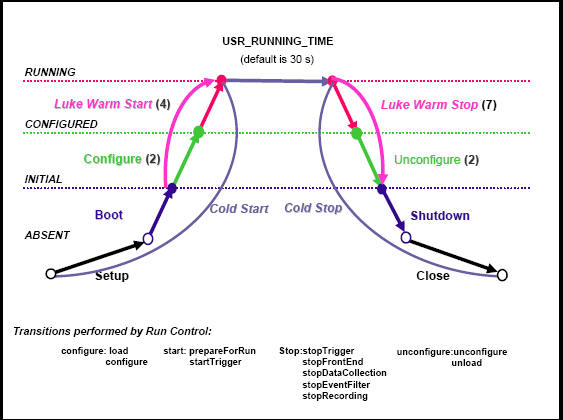
Timing tests for the state transitions:
Timing measurements on the state supervisor transitions and the rc transitions can be performed using option s in the setup_daq or play_daq scripts. Several variants are suggested
In setup_daq the option -tt or in play_daq the command line parameters time nogui will automatically run through the states as indicated in the diagram The transition times are measured. They are displayed after the shutdown is performed and printed in the results_logs_file, see setup_daq -h / play_daq -h

In setup_daq the option -ut or in play_daq the command line parameters time nogui will automatically run through the states as indicated in the diagram The transition times are measured. They are displayed after the shutdown is performed and printed in the results_logs_file ,see setup_daq -h / play_daq -h
Timing test -ut [--usercycle <.play_daq_usercycle>]
The .play_daq_usercycle is not in the release but you can
copy it from
/afs/cern.ch/user/a/atlonl/public/tools/execution/.play_daq_usercycle
The default file .play_daq_usercycle is expected to reside
in your $HOME (yes,
including the leading '.')
USR_RUNNING_TIME defines the time in running state before stopping. The default
value is 30s
Timing test -ut --usercycle <user_file>
The user can develop and use his/her own sequence of transitions. In order for results to be displayed and stored, he/she must provide the corresponding funtion. The command for setup_daq is : -ut --usercycle <path_name_to_user_timing_funtion>
The text_gui is contained in the release tdaq-01-02-00
Tests with the text_gui is a
command line gui which allows to enter commands for the state transitions. The
time is displayed after each transition and stored in the results file.
Setup_daq or play_daq can be used. The text_gui is invoked with the options
-ut --usercycle text_gui in setup_daq. The file text_gui must be copied to the
home directory or the full path to text_gui must be entered.
setup_daq_n_times:
Where can I find it ? Not yet in release tdaq-01-02-00, but you can find the script and the example described below in /afs/cern.ch/user/a/atlonl/public/tools/execution
setup_daq_n_times is a script to run setup_daq 'n' times (displayed as
loop count) for every partition listed in a partitions file, with the option of
repeating indefinitely from the start of the partition list when it has reached
the end. Read the help information given with the -h option for more info. An
example of its use is given for running over a list of partitions 20 times
each, executing a user defined timing script. Optionally a short summary mail is
send when the script is terminated. The command line would look like:
setup_daq_n_times -p myPartitionList -n 20 -ut --usercycle <pathname_of_usercycle_file>
-m <email-address>
The results files produced by a series of .play_daq_usercycle can be analysed with the tool analyse_results_usercycle3.pl.
Where can I find it ? Not yet in release tdaq-01-02-00, but you can find the script and the example described below in /afs/cern.ch/user/a/atlonl/public/tools/analysis
DESCRIPTION
Normally, using the automatic execution procedure, a set of partitions is run
n times, for example using play_daq_n_times (note that the corresponding
setup_daq_n_times is in preparation).
The analysis script produces scatter plots for each measured transition (setup,
boot, configure, etc) for each partition which is referenced in the file.
Note that the criteria is the datafile name, not the partition name.
In addition, the average of each partition for the respective number of runs
is calculated and displayed in a comparison plot per transition for all the
listed partitions, versus the number of controllers contained in the
respective partition. Failures are not counted. This is in particular useful
when having run partitions of different sizes in terms of included
controllers. The ps file can be viewed with gvv or another tool.
The analysis script produces a number of intermediate files for each partition
and for the average. The .csv files are convenient for importing the sorted data
into excel to produce presentable graphs. The .dat files are created for
each partition, for all partitions and the average of each partition. These
files do not include a header line like the .csv but rather only the timing
values.The .txt file is the file that is passed by the script to gnuplot to
create the .ps.file.
SYNOPSIS:
analyse_results_usercycle3.pl [-h] -f <input_file> -i <info_file> -o <output_file_name>
OPTIONS:
-h this help message
-f <input_file> the input file
-i <info_file> the file with the measurement info on the rc state transitions
-o <output_file_name> the file name for the global output files
EXAMPLE of analyse_results_usercycle3 USAGE, with measurements
from the tests on Westgrid:
./analyse_results_usercycle3.pl -f PLAY_DAQ_N_TIMES--meas-May19Final222---timing
-i ./timing_info_usercycle.txt -o measure19
<input_file> PLAY_DAQ_N_TIMES--meas-May19Final222---timing
<info_file> timing_info_usercycle.txt
<output_file_name> measure19
this line will generate fallowing files:
measure19.csv
measure19.dat
measure19.ps
measure19.txt
PLAY_DAQ_N_TIMES--meas-May19Final222---timing_glob.csv
Res_PARTITIONS_part_efTest.data.xml.p840_05subFarms_20nodes_1efd_4pt_0rc_0is.ParamsOmni.TimeOutS.csv
Res_PARTITIONS_part_efTest.data.xml.p840_05subFarms_20nodes_1efd_4pt_0rc_0is.ParamsOmni.TimeOutS.dat
Res_PARTITIONS_part_efTest.data.xml.p840_10subFarms_20nodes_1efd_4pt_0rc_0is.ParamsOmni.TimeOutS.csv
Res_PARTITIONS_part_efTest.data.xml.p840_10subFarms_20nodes_1efd_4pt_0rc_0is.ParamsOmni.TimeOutS.dat
Res_PARTITIONS_part_efTest.data.xml.p840_15subFarms_20nodes_1efd_4pt_0rc_0is.ParamsOmni.TimeOutS.csv
Res_PARTITIONS_part_efTest.data.xml.p840_15subFarms_20nodes_1efd_4pt_0rc_0is.ParamsOmni.TimeOutS.dat
Res_PARTITIONS_part_efTest.data.xml.p840_20subFarms_20nodes_1efd_4pt_0rc_0is.ParamsOmni.TimeOutS.csv
Res_PARTITIONS_part_efTest.data.xml.p840_20subFarms_20nodes_1efd_4pt_0rc_0is.ParamsOmni.TimeOutS.dat
Res_PARTITIONS_part_efTest.data.xml.p840_30subFarms_20nodes_1efd_4pt_0rc_0is.ParamsOmni.Timeouts.csv
Res_PARTITIONS_part_efTest.data.xml.p840_30subFarms_20nodes_1efd_4pt_0rc_0is.ParamsOmni.Timeouts.dat
Note that at the time of running ,only the root and intermediate controller was
counted and not the local controllers and therefore all partitions show 2
controllers. This has been modified for release tdaq-01-02-00 in play_daq.
For the corresponding excel graphs, an additional column has been
reated by hand containing the number of EF processes
per partition in the excel file only. It can be viewed here:
timingSeries1&2.xls
RGANG
RGANG TOOL: A SMALL INTRODUCTION (15/12/2004)
(last update:06/06/2005)
TOOL TO EXECUTE COMMANDS OR SCRIPTS IN SEVERAL MACHINES IN A PARALELL WAY
[Note: The following commands and scripts have not yet been tested in more than
20 machines in paralell in the testbeds
Still waiting for doing those tests when machines are available]
IMPORTANT NOTES:
The file rgang.py, as well as the following described scripts, are under
These files should be in the PATH in order to work properly.
They can be found together with rgang.py under /afs/cern.ch/user/s/sobreira/public.
INSTRUCTIONS:
RGANG options (switch):
--nway=1 ----> done in 'serial' mode
--nway=200 (BY DEFAULT) ----> very fast (more machines executing in
paralell-maximun number)
A typical RGANG command has the following sintax:
rgang.py farmlet_name 'command arg1 arg2 ... argn'
--> farmlet_name manually entered:
Range of machines: rgang.py "pcatb{132-140}" 'command arg1 arg2 ... argn'
Set of machines: rgang.py "pcatb{132,140}" 'command arg1 arg2 ... argn'
--> farmlet_name can also be a file, containing the machines' names:
#Comment
pcatb132
pcatb133
pcatb134
pcatb135
pcatb136
pcatb137
pcatb138
pcatb139
pcatb140
RGANG can be very useful for performing commands in several machines (executing
in paralell):
typical commands performed may be:
> Copying files to several nodes (rgang's copy mode - take care of the -c switch
after rgang):
rgang.py -c "pcatb{132-140}" origin_file destination_file
Example: rgang.py -c "pcatb{132-140}" /tmp/test_orig /tmp/test_dest
.Check if the file has really (it should!!) been copied: rgang.py -x
"pcatb{132-140}" ls /tmp/test_*
(without -c switch, since we don't want to copy anything-copy mode not required)
.Remove the file: rgang.py -x "pcatb{132-140}" rm /tmp/test_dest
> Executing files or commands in several nodes (not in the copy mode, no -c
switch)
The -x switch avoids some "annoying" messages
rgang.py -x "pcatb{131-140}" path_file_to_execute
Examples:
rgang.py -x "pcatb{132-140}" rm /tmp/test_dest
rgang.py -x "pcatb{132-140} ~/scripts/testscript --->Range of machines
rgang.py -x "pcatb{132,137,139}" ~/scripts/testscript --->Set of machines
rgang.py -x pcatb{131-140} ls /tmp
rgang.py -x "pcatb{132-140}" 'ls /tmp | grep test_dest'
USEFUL COMMAND:
rgang.py -x "pcatb{131-140"} ps -u username ---> displays all processes
belonging to the user username on the given set of nodes
> Measuring command execution performance:
time rgang.py -x --nway=4 pcatb{131-140} /home/sobreira/rgang/bin/script --->
slow
time rgang.py -x --nway=10 pcatb{131-140} /home/sobreira/rgang/bin/script --->
faster (done more in paralell)
time rgang.py -x pcatb{131-140} /home/sobreira/rgang/bin/script ---> much faster
(default nway=200)
###########################################SCRIPTS DEVELOPED USING/FOR (ON TOP
OF) RGANG###########################################
> "show" script
it can be performed using rgang (several machines in paralell) or in just one
machine (regular usage)
The script can:
-h option: display help and some examples
#---------------------------------INCLUDE USER
Section----------------------------------#
-u option: show all processes belonging to user
Examples:
#RGANG USAGE: rgang.py -x pcatb{131-140} show -u "werner\ sobreira\ root"
#REGULAR USAGE: show -u werner\ sobreira\ root
#---------------------------------EXCLUDE USER
Section----------------------------------#
-v option: show all users and their processes, except those refering to the
speficied users
Examples:
#RGANG USAGE: rgang.py -x pcatb{131-140} show -v "werner\ root\ sobreira"
#REGULAR USAGE: show -v werner\ root\ sobreira
#--------------------------------INCLUDE PATH
Section-----------------------------------#
-p option: show all users performing the processes refered by their path
Examples:
#RGANG USAGE: rgang.py -x pcatb{131-140} show -p "/sbin\ pmg\ /usr\ /usr/libexec/post\
/afs/cern.ch/"
#REGULAR USAGE: show -p /sbin\ pmg\ /usr\ /usr/libexec/post
#--------------------------------EXCLUDE PATH
Section-----------------------------------#
-q option: show all users performing whichever processes except those which are
specified
Examples:
#RGANG USAGE: rgang.py -x pcatb{131-140} show -q "pmg\ /afs\ /sbin\ /usr\ crond\
ypbind\ /afs"
#REGULAR USAGE: show -q pmg\ /afs\ /sbin\ /usr\ crond\ ypbind
NOTE: Just the beginning of the arguments (users or paths) might be specified
and multiple arguments (users or paths) should be separated by "\ " (slash and
space)
###################
> "collect" script
It can be only be performed using rgang utility.
One run of the script "overlaps" a previous run, so that previous a node's name
directory can be deleted if
that node doesn't contain any searched file
NOTE: * must be preceed by an \ when specifying more than one file
"Collect" certain file(s) from several nodes and send a copy to a specified node
in a specified directory
Files are kept under a directory with the name being the provenience node name
Syntax: rgang.py -x farmlet_name collect path_file
destination_node:directory_to_place_files
Examples: rgang.py -x pcatb{131-140} collect "/tmp/expfile\*" $HOST:~/exp
(collecting all files in the tmp directory starting by 'expfile' and placing it
in the actual machine
in a directory called exp)
rgang.py -x pcatb{131-140} collect "/tmp/expfile\*" pcatb135:/tmp
(collecting all files in the tmp directory starting by 'expfile' and placing it
in the pcatb135 machine
in the 'tmp' directory under the 'pcatb135' directory)
rgang.py -x pcatb{131-140} collect "/tmp/expfile4.teste" $HOST:~/exp
(collecting a file called /tmp/expfile.test and place it in ~/exp/pcatbxx
##################
> "machines_up" script
It uses rgang internally; therefore, rgang.py must be in the PATH in order to
perform this script
Shows machines which machines are up in a given set of nodes and stores then in
a given file, so that
this file can be used to specify instructions using rgang
Syntax: machines_up -s set_of_machines_to_analyze [-f file_to_store_machines_up]
Examples:
machines_up -h ---->(help) to display the usage
machines_up -s "pcatb{131-180}" -f /tmp/up
machines_up -s "pcatb{131,135,152,180}" -f ~/up_machines
Monitoring system gives status of nodes, CPU
consumption and network traffic,
see
http://ccs003d.cern.ch/lemon-status/
the Quattor system manages the nodes for sw
installation, repository updates,
servers and daemons to run, etc according to an installation
list
Monitoring of the farm (to be confirmed)
-----------------------
The subset of nodes that will be assigned to us should be available in
ythe monitoring system as Atlas-tdaq.
to be made available:
- a file containing the host names of the nominal list of nodes
belonging to Atlas-tdaq, one per line.
- a file containing the host names of the actual list of nodes
belonging to Atlas-tdaq, one per line.
'actual' means that the nodes is in a state in which it
responds to
ssh. It is important that this file is automatically updated
when a
change in actual availability occurs.
copies originalPartitionName to newPartitionName